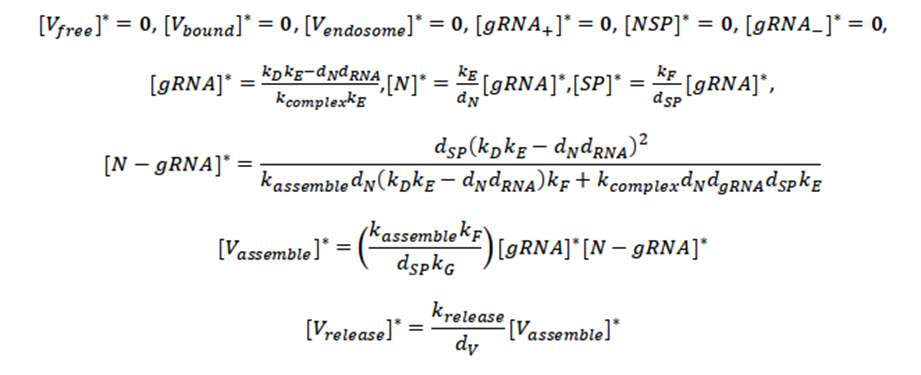First Analytical Solution of Intracellular life Cycle of SARS-CoV-2
Abstract
The goal of this paper is to show that it is possible to obtainan analytical solution of the life cycle of SARS-CoV-2 based on a deterministic model. To do this, this work solved a system of twelve differential equations where we obtained two points of equilibrium. The first critical point corresponds to the initial conditions regarding the virus entry into the cell without replication in the cell, and the second involved one is the process of the transcription and replication of the virus in the infected cell.
Author Contributions
Copyright © 2022 Raúl Isea, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
The new Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV-2) is a virus belonging to the Coronaviridae family in the order nidoviralesand subfamily Coronavirinae1. The disease it causes is known as Coronavirus disease 2019 (COVID-19), and was declared a pandemic on March 11, 2020. To date, there have been around 450 million cases of Covid-19 until March 6, 2022 according to the information obtained from the Universidad Johns Hopkins (available at coronavirus.jhu.edu).
It is a virus of approximately 28-32 kb. It is a 5’-capped and 3’-polyadenylated positive-sense single strand RNA (+ssRNA), non-segmented, and belongs to the genus of betacoronaviruses2, similar to the structure found in messenger RNA (mRNA) of eukaryotic cells 1, 2.
The replication of the virus begins when the S protein of SARS-CoV-2 binds directly to the Angiotensin-Converting Enzyme 2 (ACE2) receptor 3. It is interesting to note that ACE2 receptors are expressed in several organs of the human body (i.e., lungs, kidneys, intestine), with an affinity 10–20 times higher than with SARS-CoV 4.
Once the virus has entered the host cell, it is released into the cytoplasm, starting the replication process that will give rise to non-structural proteins and also accessory proteins, with four structural proteins 5. The formation of RNA(-) and also replication and transcription of Rna subgenomics 6. These subgenomic RNAs(-) are transcribed into mRNAs(+) which encode the structural proteins S, M, E, N, and accessory proteins 7. During the replication process, the N protein of the virus binds to the genome, while the M protein associates with the membranes of the endoplasmic reticulum (ER). Finally, the virions are secreted by exocytosis 3, 4.
Taking into account the above description, the next step is to generate a mathematical model of the life cycle of SARS-CoV-2 using the law of mass actionas described in the next session. This type of study has been used in other intracellular replication processes such as HIV-1 8, hepatitis B virus 9, influenza A virus 10, among others.
Mathematical Model
There are many scientific studies about the life cycle of SARS-CoV-2 (see for example 3, 4, 5, 6), but only one is based on differential equations that describe the previous process led by Dmitri Grebennikov et al 11. They proposed a system of twelve differential equations which they solved numerically and split their model in (1) cell entry, (2) genome transcription and replication, (3) translation of structural and accessory proteins, and finally (4) assembly and release of virions. Based on this model, we are going to changes some of these equations using the same Grebennikov’ definition, as explained below.
Cell Entry
The equations that describe the process of the cell entry are the same ones used by Grebennikov et al 7.There are four differential equations, where the free virions outside of the cell ([Vfree]) will be binding the ACE2, and activated by TMPRSS2 ([Vbound]), while that [Vendsome], and [gRNA+] are the number of virions in endosomes and the number of ss-positive sense genomic RNA, respectively. The equations are
where the value of the constants are indicated in Table 1.
Transcripción y Replicación Del Genoma.
The nonstructural proteins (Nsps) are responsible for the transcription and replication of genomic RNA, where Nsp12 is the central component in encoding the RNA-dependent RNA polymerase (RdRp), and its function is to generate a negative-sense single-stranded RNAs.
In this step, Grebennikov’s model proposes three differential equations, but we modified it (the changes are indicated in red color), where the first equation, NSP, takes into account the abundance of nonstructural protein populations, while that [gRNA-] takes account the negative sense genomic and subgenomic. Finally, the last equation is completely different from Grebennikov's model which describes [gRNA]
(Remember that the values of the constants are indicated in Table 1).
Table 1. Model parameters based on Grebennikov‘s model 11, with the exception of some parameters indicated in red color.| Parameter | Value |
| K diss | 0,61 |
| K bind | 12,00 |
| K fuse | 0,50 |
| K uncoat | 0,50 |
| K complex | 0,40 |
| K assemble | 1,00 |
| K release | 8,00 |
| dn gRNA | 0,20 |
| d NSP | 0,07 |
| dn gRNA _ | 0,10 |
| d N | 0,02 |
| d SP | 0,04 |
| d N - gRNA | 0,20 |
| d N - gRNA | 0,12 |
| d endosome | 0,06 |
| K A | 2,10* |
| K B | 2,99* |
| K C | 0,20* |
| K D | 0,19* |
| K E | 37,32* |
| K F | 4,39* |
| K G | 8,01* |
Traducción de Proteínas Estructurales y Accesorias.
We are going to consider very simple equations that describe the number of N proteins per virion (N) and the total number of structural proteins (SP) unlike Grebennikov, where we are not going to consider the formation of virus-like particles and the budding of new virions from the ER and Golgi compartments (ERIGC), so the equations in this model are:
Finally, the Assembly and Release of Virions
The N proteins play an important role in incorporating viral RNA into particles. The virions assemble in the ER-Golgi compartment by encapsulation of N-RNA complexes and the newly assembled virions can leave the infected cell by exocytosis.
To explain this step, we change the equation that describesthe rates of change of the ribonucleocapsid (N), while the assembled virions ([Vassemble]), and released ([Vrelease]) are described as:
The next step is solving these equations analytically.
Results and Discussions
This system of differential equations is solved employing the same methodology used and validated in previous works 10, 12, 13, 14, where the epidemiological relevant region (W) is given by
The next step is to find the critical point. We find two critical points to be described below:
First Critical Point:
The trivial solution of the system of differential equations occurs when they are all equal to zero:
It means that there are no free virions and neither infected virus (all values are zero).
The next step is to calculate the Jacobian of the system and evaluate it at this critical point. We only showthe non-zero terms of the Jacobian (J), where the order of the subscripts correspond to the row, column of the Jacobian matrix, respectively; that is, J4,3 corresponds to the value of the matrix at row position 4, column 3 of the resulting Jacobian matrix. Thus, all non-zero terms are
Where k1,k2 and k3 as (kbind+dv),(kfuse+kdiss+dv) and (kuncoat+dendsome) respectively;
From these expressions, four eigenvalues of the system are obtained, which indicate the stable conditions of the system, given by:
It is interesting to note that there is no restriction for the system to be stable since the expression in the square root will always be positive. Likewise, it is surprising that the stability of the system depends precisely on the variables of the initial contagion of the virus, that is, on the entry of the virus.
Second Critical Point
This critical point reveals that the virus can be transcription and replication without being present in the environment.
Finally, we are going to determine if these equilibrium points are stable, and to do that, we calculated the eigenvalues for this critical point, but they are not indicated in the work due to the complexity of the equations, so the five eigenvalues for the second critical point are indicated, which are:
The first two eigenvalues coincide with those of the first critical point, while the last equations essentially depends on the virus release mechanism.
Numerical Evaluation
To demonstrate the feasibility of the model, the system of differential equations was solved using a program written in Python, where the initial values are 10,10,2,4,0,10,10000,456,2000,0,0,1 which correspond to [Vfree], [Vbound], [Vendsome], [gRNA+], [gRNA] [NSP], [gRNA_] , [N],[SP], [N-gRNA], [Vassemble] and [Vrelease] respectively.
The results are shown in figure 1, Figure 2, and Figure 3. Figure 1 corresponds to the entry of the virus into the cell and logically, when using these equations with the values of the constant, said trend coincides with respect to the work of Grebennikov et al.
Figure 1.The free virions Vfree decreases as binding to ACE2, and are generated gRNA(+).
Figure 2.It is interesting to note that there is a production of N over time even though it decreases gRNA_.
Figure 3.Virus particles released from the cell.
Figure 2 begins to differ from the model proposed by Grebennikov’s model basically the dynamics of N and SP, and for this reason, it is necessary to adjust the parameters with experimental values to know how valid is this model proposed in this paper. Finally, figure 3 shows the release process of the particles over time with the same trend as that presented by Grebennikov, but the release time is shorter in this work, unlike the one presented previously. Therefore, further studies of the model constants should be carried out in order to determine theiraccuracy.
Finally, when numerically calculating the eigenvalues according to the data indicated in Table 1, three of the four eigenvalues are negative (-12.66; -0.59; -2.77) corresponding to the first critical point, which indicates that this system is stable. In the case of the second critical point, all five eigenvalues are negative, that is, -354.54; -0.02; -12.66; -0.59; -2.30, and also it is stable the system.
Conclusions
The present paper shows that it is possible to analytically resolve the SARS-CoV-2 life cycle. In fact, we obtained two conditions of equilibrium, that is, when the virus is present in the environment, and when it is transcribed and replicated in the infected cell. Finally, more studies must be done to obtain the correct values of the constants indicated in the differential equations and from there, to be able to carry out the stability studies.
References
- 1.Yang Y, Xiao Z, Ye K, He X, Sun B et al. (2020) SARS-CoV-2: characteristics and current advances in research.Virology. Journal,17: 117.
- 2.Pal M, Berhanu G, Desalegn C, Kandi V. (2020) . Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): An Update.Cureus12(3):e7423 .
- 3.Jackson C B, Farzan M, Chen B, Choe H. (2021) Mechanisms of SARS-CoV-2 entry into cells.Nature Reviews Molecular Cell Biology,23:. 3-20.
- 4.Cao Y C, Deng Q X, Dai S X. (2020) Remdesivir for severe acute respiratory síndrome coronavirus 2 causing COVID-19: An evaluation of the evidence.Travel Med Infect Dis. 101647.
- 5.Kim Y, Jedrzejczak R, Maltseva N I, Wilamowski M, Endres M et al. (2020) . Crystal structure of Nsp15 endoribonucleasaNendoU from SARS-CoV-2.Protein Sci 29, 1596-1605.
- 6.Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. (2020) A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping.Cells. 9(5), 1267.
- 7.Tang T, Bidon M, Jaimes J A, Whittaker G R, Daniel S. (2020) Coronavirus membrane fusion mechanism offers a potential target for antiviral development.Antiviral Res (Internet). 178, 104792.
- 8.Schberbatova O, Grebennikov D, Sazonov I, Meyerhans A, Bocharov G. (2020) Modeling of the HIV-1 life cycle in productively infected cells to predict novel therapeutics targets.Pathogens9:. 255.
- 9.Fatehi F, Bingham R J, Dykeman E C, Patel N, Stockley P G et al. (2020) An intercellular model of Hepatitis B viral infection: an in silico platform for comparing therapeutics strategies.Viruses13:. 11.
- 10.Isea R. (2018) Analytical Solutions for the Initial Steps of the Intracellular Dynamics of Influenza A Virus.Acta Scientific. , Microbiology 1, 6-8.
- 11.Grebennikov D, Kholodareva E, Sazonov I, Karsonova A, Meyerhans A et al. (2021) . Intracellular Life Cycle Kinetics of SARS-CoV-2 Predicted Using Mathematical Modelling.Viruses13: 1735.
- 12.Isea R. (2016) A Preliminary Mathematical Model for the Dynamic Transmission of Dengue. , Chikungunya and Zika.American Journal of Modern Physics and Application 3(2), 11.
Cited by (3)
This article has been cited by 3 scholarly works according to:
Citing Articles:
R. Isea - Journal of Model Based Research (2023) Semantic Scholar